The Crino Laboratory
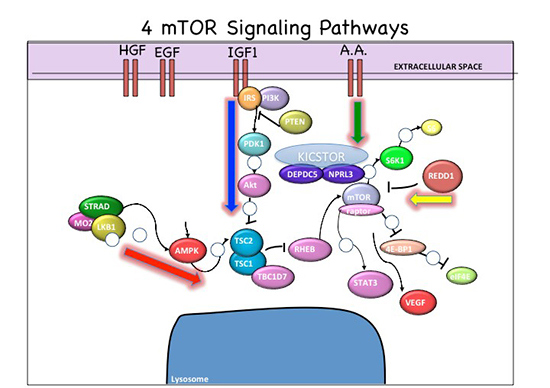
Signaling Pathway Vectors within mTOR Cascade. Red arrow, energy pathway; blue arrow, growth factor pathway, green arrow, amino acid pathway, yellow arrow, oxygen sensor pathway.
My lab has maintained an NIH-funded translational research program studying developmental brain malformations linked to epilepsy, autism, and intellectual disabilities for the past 20 years.
We have specific expertise in defining developmental disorders associated with aberrant mammalian target of rapamycin (mTOR) signaling and we were the first lab to define altered mTOR signaling in focal cortical dysplasias.
Our efforts led to the nomenclature to define the family of mTOR associated disorders ("mTORopathies"; see Crino, 2011) including focal cortical dysplasia (FCD), hemimegalencephaly, ganglioglioma, and Pretzel syndrome (PS).
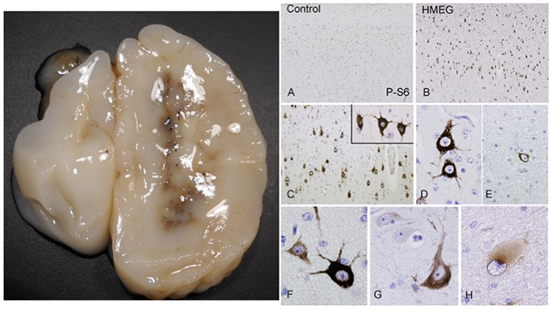
Hemimegalencephaly. Left, note massive enlargement of hemisphere. Right, numerous dysmorphic neurons exhibiting hyperphosphorylation of ribosomal S6 protein.
A major focus of my lab over the past two decades has been tuberous sclerosis complex (TSC) in which we have worked on human brain tissue biomarkers as well as mouse and in vitro models.
My lab has successfully implemented numerous experimental strategies in resected human tissues including immunohistochemistry, stereology, Western analysis, mRNA expression analysis, and DNA mutation analysis.
We have successfully implemented cell culture model systems to study the effects of mTOR genes such as Tsc1, Tsc2, and STRADA, in vitro models on cell development using immunocytochemistry, Western assay, mRNA expression analysis, gene transfection, and in vitro cell migration assays. In addition, we have optimized and implemented strategies in fetal mouse cortex using in utero electroporation to study the cell lineage marker, cell size, and cell signaling assays.
We have extensive experience testing pharmacological compounds such as the mTOR pathway inhibitors in cell culture and live animals. International collaborations have helped to describe mutations in GATOR1 complex proteins (NPRL3 and DEPDC5) in FCD (Scerri et al., 2015; Sim et al., 2015). Most recently, we were part of a large collaborative initiative to define genes responsible for non-lesional neocortical epilepsy (Winawer et al., 2018).
View Dr. Crino's University of Maryland School of Medicine profile.
Pretzel Syndrome: A Rare mTORopathy
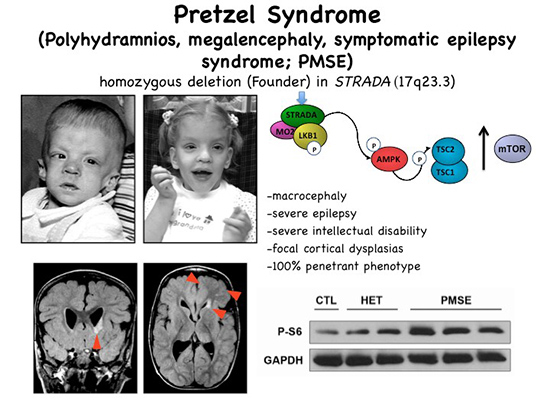
Pretzel syndrome resulting from STRADA mutation leads to hyperactivation of mTOR signaling. Note enlarged head size in 2 children. Brain MRI reveals subependymal heterotopia and area of FCD. Bottom right, hyperphosphorylation of S6 protein in patient (PMSE) versus heterozygous (unaffected parents) and control fibroblasts.
In 2004, the Crino Laboratory began what would become a 14 year collaboration with the Clinic for Special Children (CSC) in Lancaster, PA to investigate the genetic causes of Pretzel syndrome (PS), a rare autosomal recessive, neurodevelopmental disorder associated with severe, pediatric onset, intractable epilepsy, profound intellectual disability, megalencephaly, and focal cortical dysplasia.
In 2007, we collaboratively helped to demonstrate that all PS patients share a common homozygous truncating deletion of exons 9-13 in the STE20-related kinase adaptor alpha (STRADA) gene, removing the protein interacting domains for complex formation with the liver kinase B1 (LKB1), a known upstream mTOR pathway gene.
Our lab then embarked on a collaborative investigation of the role of STRADA in brain development showing a clear link via AMPK to mTOR regulation, working in vitro and making a mouse model, showing that loss of STRADA led to exuberant hyperactivation of mTOR. In further studies, we developed new experimental validation techniques to demonstrate definitive effects of STRADA on cell size, polarity, and motility, cortical lamination, brain size, and neuronal hyperexcitability.
Subsequent NIH funded collaborations have yielded iPSC-derived neurons and a knockout mouse. Based on all of our pre-clinical data to functionally validate STRADA as a major mTOR regulator, we treated 6 Mennonite PS children (open label) with rapamycin prior to the onset of seizures, reasoning that the uncomplicated genetic background of the Mennonite community, a recessive inheritance, and a Founder mutation would provide a unique opportunity to test whether mTOR inhibition prior to seizure onset might alter the clinical course of these patients.
Our decade-long work on STRADA in PS demonstrates our abilities to proceed from gene identification to functional protein validation using a number of lab-developed assays, to successful proof-of-principle clinical trial in human patients.
Crino Laboratory Staff
Marianna Baybis, MS – Lab Manager
Janice K. Babus, BS – Research Specialist
Soad Elziny, BS
Ms. Elziny Is a Ph.D candidate in the Program in Neuroscience at the University of Maryland, Baltimore. She received her BS in Neuroscience from Virginia Tech. Her research focuses on defining disease mechanisms in FCD 1A and non-lesional epilepsies.
Techniques Used in the Crino Lab
- Immunohistochemistry
- Immunocytochemistry
- Quantitative Cell Morphometry
- Confocal Microscopy
- PCR, RT-PCR
- Gene cloning
- CRISPR/Cas9 gene editing
- shRNA Knockdown
- Plasmid overexpression
- Mouse modeling
- In utero electroporation
- Stereotaxic microinjection
- FRET and live-cell imaging
Recent Publications
- Somatic SLC35A2 variants in the brain are associated with intractable neocortical epilepsy
- DEPDC5 and NPRL3 modulate cell size, filopodial outgrowth, and localization of mTOR in neural progenitor cells and neurons
- Tuberous sclerosis complex
- Homozygous boricua TBCK mutation causes neurodegeneration and aberrant autophagy
- Somatic uniparental disomy of Chromosome 16p in hemimegalencephaly
- Coding and small non-coding transcriptional landscape of tuberous sclerosis complex cortical tubers: implications for pathophysiology and treatment
- Focal Cortical Dysplasia: Gene Mutations, Cell Signaling, and Therapeutic Implications
- Genetics of tuberous sclerosis complex: implications for clinical practice
- The mTOR signaling cascade: paving new roads to cure neurological disease
- Sending Mixed Signals: The Expanding Role of Molecular Cascade Mutations in Malformations of Cortical Development and Epilepsy
- Novel Histopathological Patterns in Cortical Tubers of Epilepsy Surgery Patients with Tuberous Sclerosis Complex
Positions Available
Post-doctoral fellows, PhD students, technicians, and undergraduates interested in working in our lab on developmental brain disorders should contact Dr. Crino at pcrino@som.umaryland.edu or Marianna Baybis at mbaybis@som.umaryland.edu
